One of the important applications of ion beam irradiation is in biology, e.g. for cancer therapy and crop mutation breeding. In the applications, the irradiation finally modifies or changes DNA in the tissue cells so that cell death or mutation can be triggered. In the irradiation of materials, the particle energy deposition in the target atoms dominantly occurs around the Bragg peak which is very near the end point of the ion traveling in the material. At this stage, the ion energy becomes very low because of the energy loss in the ion traveling path. Therefore, ion irradiation of DNA at the very low energy regime is of particular research interest in understanding of fundamentals involved in the interaction between the ultra-low-energy ions and DNA. In the interaction, effect on DNA double strand breaks (DSBs) has special significance due to the fact that DSBs can hardly be repaired or correctly repaired, consequently leading to cell mutation or death. In studying ion radiation effects in DNA, computer simulations, especially with powerful software, have been recently initiated. The most recently developed software for the simulation of ionizing radiation of DNA is Geant4-DNA. Geant4-DNA is an extension from the well-known Geant4 which is a general–purpose Monte Carlo simulation toolkit for the simulation of the passage of particles through matter (developed by CERN). The main objective of the software Geant4–DNA is to simulate biological damages induced by ionizing radiations at the cellular and sub–cellular scale (developed by European Space Agency). Although there has been some work reported on using Geant4-DNA mostly to study light particle irradiation effects on water, there have been rare investigations of using the software package for ultra-low-energy (< 100 eV/amu) ion irradiation of naked DNA, particularly focused on DNA SSBs and DSBs. In the most recent public release of Geant4 (version 9.3 BETA, released in June 2009), Geant4-DNA physics processes are applicable to particles down to an energy limit of 8.23 eV for electrons, corresponding to the lowest excitation level of the liquid water molecule. Protons and hydrogen are tracked down to 100 eV, while helium particles and their charged states down to 1 keV. The particle energy that we applied in the simulation was around the low limits of the applicable energy ranges in Geant4-DNA and this was our special interest.
The Geant4 version 10.01.02 on the basis of the pdb4dna example was used to study the particle-irradiation – DNA interaction. Fig. 1 illustrates the computational steps. The first step was generating our own modified Protein Data Bank (PDB) files to model DNA by reading HEADER which gave the type of DNA, ATOM which provided the information and coordinates of each atom, and TER which indicated the end of the file. The generated file was inserted into the Monte Carlo (MC) code to simulate the interactions. Occurrences of SSBs and DSBs were simulated with different DNA models, particle types, energy, and dose/fluence. Interactions between particle irradiation and DNA included ionization, excitation, charged transfer, and indirect actions of radiations, e.g. secondary high/low energy ions and electrons generated, and hydroxyl radicals [1]. It must be noted that these interaction processes are originally set up by Geant4-DNA which claims these processes dominant, though in our opinion, direct atomic displacement due to elastic collision between the particles and the DNA atoms should be included. Three models of 30–base–pair–long A–form DNA duplex, i.e. poly–AT, consisting of only adenine and thymine bases, poly–CG, consisting of only cytosine and guanine bases, and pGFP, containing the green fluorescence protein, in the atomistic geometries of the DNA duplex with sequence 5′-AAGAATGGAA TCAAAGTTAA CTTCAAAATT–3′), were constructed for investigation. Fig. 2 shows the three modeled DNA duplexes constructed by the program. Limited by availability from the source data of the currently downloaded and installed software, light particles including electron, hydrogen, proton (H+), alpha (He2+), alpha+ (He+) and helium (neutral He) were applied for irradiation. The particle energy was varied at 10, 100, 500, 750 and 1000 eV and the particle beam dose or fluence was varied as 5 × 103, 1 × 104, 5 × 104 and 1 × 105 particles/cm2. All these conditions of the particle beam as well as the DNA characters such as the distance between base pairs could be configured in runInGUI.mac file. In the simulation, the calculation volume that contained the DNA duplex was irradiated isotopically by the particles coming from all directions, as shown in Fig. 3, in principle, different from conventional particle beam irradiation where the particles come from only one direction. The simulation-generated results were analyzed and summarized from the PDB4DNA which displayed the entries representing the actual quantity of the irradiating particles, the deposited energy and the numbers of SSBs and DSBs, as illustrated in Fig. 4.
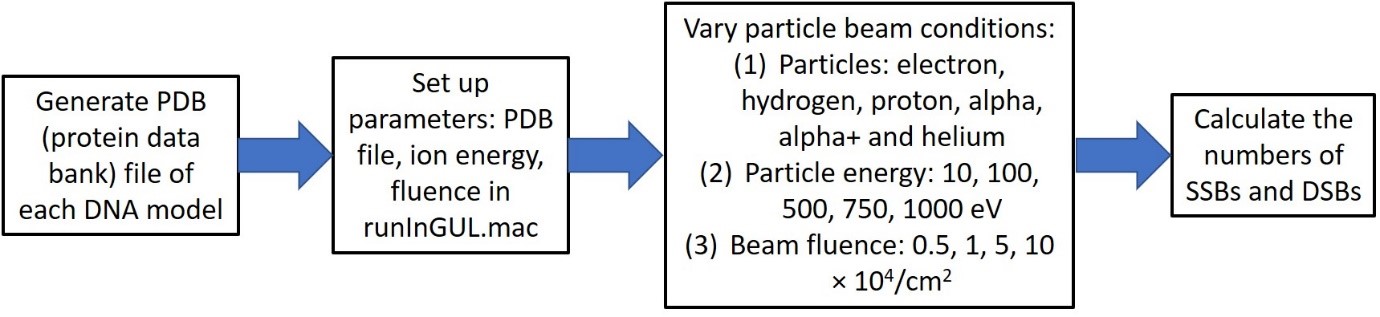
Fig. 1. Frame diagram of the computational steps of using Geant4-DNA.
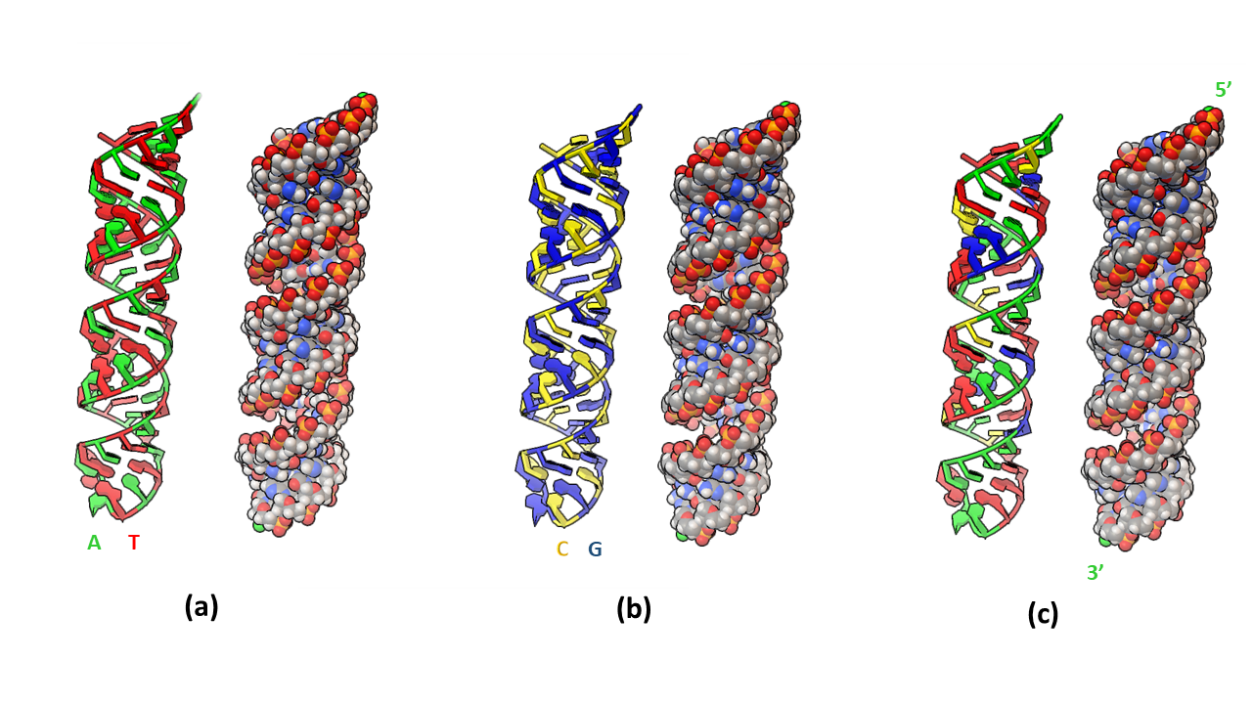
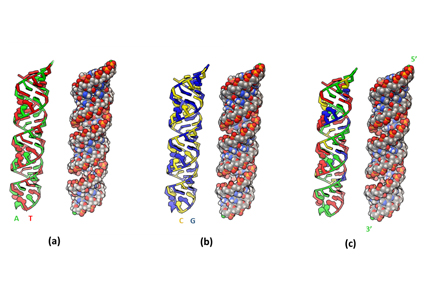
Fig. 2. Modeled three A-form DNA duplexes: (a) poly-AT, (b) poly-CG, and (c) pGFP. In each model, atoms are represented as spheres on the right with color coding: white – hydrogen, grey – carbon, red – oxygen, blue – nitrogen, and orange – phosphorus.
Fig. 3. A program display of the practical Geant4-DNA simulation of particle irradiation of DNA target, showing particles isotopically irradiating the target DNA. The straight lines represent the particle incidences and the bright spots represent DNA atoms.
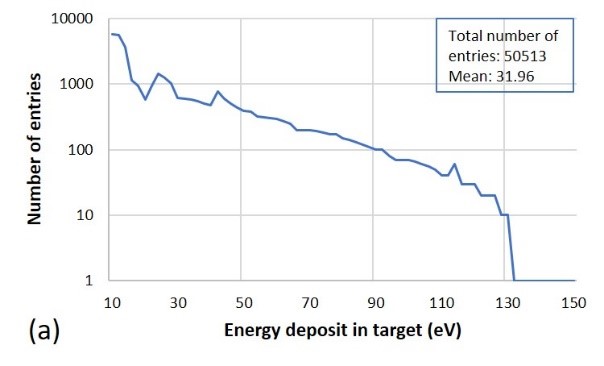
Fig. 4. Output of calculated results from irradiation of poly-AT by 100-eV electrons at a beam fluence of 1 × 105 particles/cm2, including (a) the deposited energy distribution, and (b) the numbers of SSBs and DSBs. The DNA strand closeness index: the lower the number, the closer the strands.
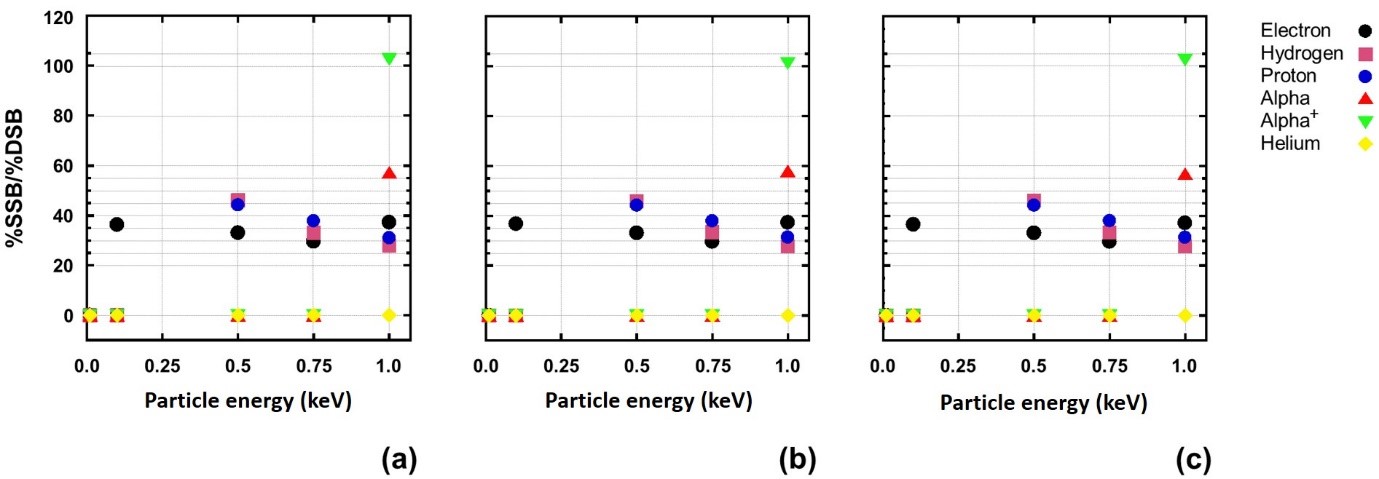
Fig. 5. Example of the SSBs/DSBs ratio as a function of the particle energy from (a) poly-AT, (b) poly-CG, and (c) pGFP at a particle fluence of 105 particles/cm2.
From the example shown in Fig. 4, it is observed that the particle energy deposition mostly takes place at the particle energy as low as possible, demonstrating the Bragg peak effect; SSBs are considerably more than DSBs by near 40 times (the number ratio of entries between SSBs and DSBs is exactly 5403/149 = 36.3, in the shown case); and DNA strand breaks, particularly DSBs, most probably happen at two most adjacent sites of the double strands. Figs. 5 and 6 summarize the results on SSBs and DSBs as functions of the particle energy and the dose for all kinds of the ionizing particles and all DNA models used in the simulation. From Fig. 5, one can directly read the ratios of SSBs/DSBs which are mostly around 36. From Fig. 6, the SSBs/DSBs ratios which can be obtained from the SSB values shown in (a), (b) and (c) divided by the DSB values shown in (d), (e) and (f) for the three DNA models, respectively, are also seen mostly around 36. This particular ratio of SSBs/DSBs may be understood from a graphic illustration (refer to our paper, as shown in Fig. 7).
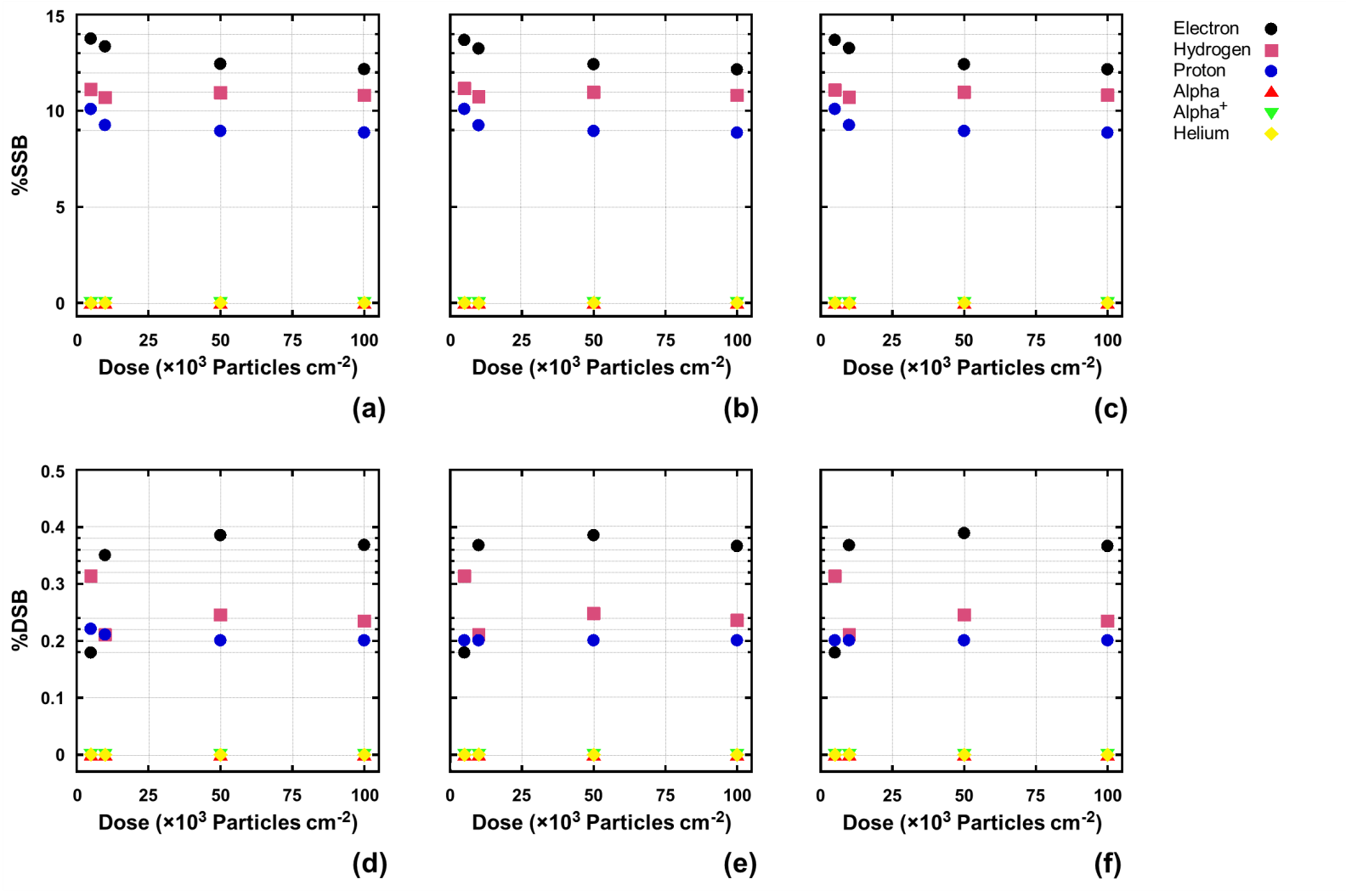
Fig. 6. Examples of SSBs [(a), (b) and (c)] and DSBs [(d), (e) and (f)] in percentage as a function of the particle fluence (dose) from poly-AT [(a) and (d)], poly-CG [(b) and (e)], and pGFP [(c) and (f)] at particle energy of 500 eV.
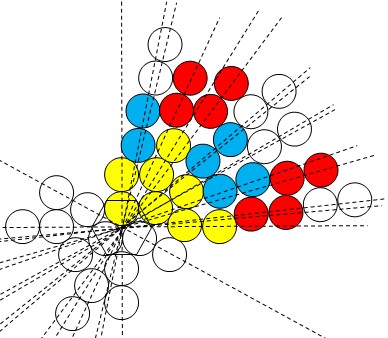
Fig. 7. Graphic illustration of the probability of occurrence of particle-irradiation caused DNA DBSs. Particle start from the origin to irradiate DNA double strands at random. A pair of balls along the irradiating direction (represented by dash lines) represent the most adjacent two atoms in the double strands, respectively. Yellow atoms are primarily impinged (directly by the particle), blue atoms secondarily impinged (the particle has to pass mostly through a yellow ball before impinging them), and red atoms tertiarily impinged (the particle has to pass mostly through two balls before impinging them), while other atoms in white (in the 1st quadrant) impinged in “higher orders” (the particle has to pass through more than three balls before impinging them). (For a detailed explanation, please refer to our paper.)
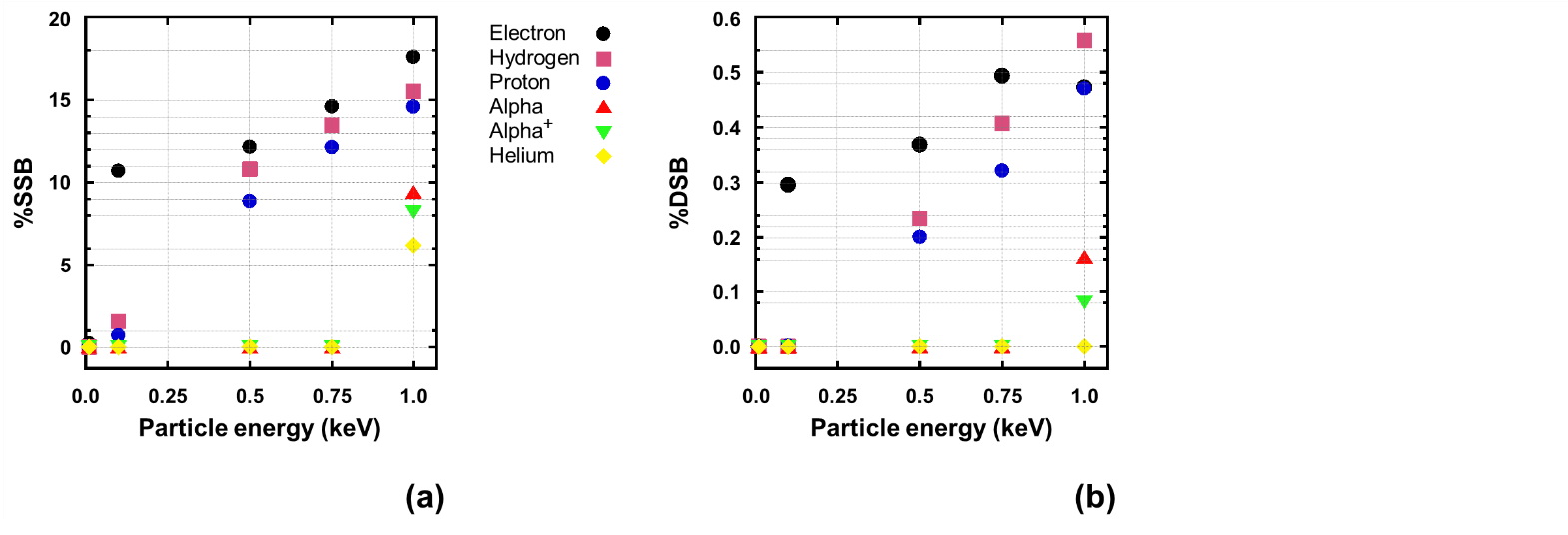
Fig. 8. An example of the particle energy dependence of DNA strand breaks. (a) SSBs. (b) DSBs. DNA model: poly-AT. Particle beam fluence: 105/cm2.
The DNA strand breaks are seen dependent on the particle energy and mass, as shown in Fig. 8. Both SSBs and DSBs increase obviously with increasing of the particle energy. There exist particle energy thresholds for DSBs to occur. At extremely low energy no DSBs occur. Energy threshold for DSB using electron was observed around 100 eV which is the lowest energy threshold case in this study, following by hydrogen and proton at 500 eV. The highest energy applied in this study at 1000 eV can introduce DSB using alpha and alpha+ but not for helium. This result is in principle in agreement with our previously obtained experimental evidence, indicating the lighter particles more effective in causing DNA change [3]. Fig. 8 also displays the dependence of the SSB and DSB fractions on the irradiating particle mass in such a general trend that the lighter the particles, the higher the strand break percent fractions for both SSBs and DSBs. This seems contradictory to common understanding on the ion–solid interaction. At the same energy for the same target material, a heavy ion (or particle) has higher linear energy transfer (LET) than a lighter ion (or particle) because of its more stopping (S) and thus deposits energy in a higher intensity, leading to more effective radiation damaging to the target. The simulation actually shows DSBs critically related to irradiating particle’s velocity, namely, the higher the velocity, the more the DSBs. Because the particle velocity is associated to ionization, here we may refer the result to such a speculation that ionization is focused by Geant4-DNA to be the dominant physical interaction process, whereas atomic displacement due to incident particle knocking out the target atoms is overlooked. We would here further point out that not only elastic scattering but also elastic collision caused atomic displacement should be definitely included in the simulation toolkit, particularly for heavier ions.
No were observable differences in inducing the strand breaks shown among different DNA models. This might imply that in the software, the constructions of different DNA models applied the same parameters for the DNA atoms and bonds. This result seemed different from that obtained by molecular dynamics simulation, which showed a preferential effect of ion irradiation of DNA on DNA changes [4], in more agreement with experimental results [5], e.g. A-T more vulnerable to G-C [6]. As for the particle charge effect, it showed somewhat complexity. In a general concept, charged particle irradiation should have more effect than neutral particle irradiation on modification of target materials owing to the additional Coulomb interaction, which is proportional to the quantity of charge. Therefore, for the case of charged and uncharged helium irradiations, the result is very reasonable, whereas for the case of hydrogen and proton irradiations, it is currently hardly understood yet. We here propose a hypothesis which refers to the difference in the traveling distance between a neutral and an ion for an interpretation (for details, please refer to our paper).
In conclusion, the simulation showed some agreements and also some discrepancies between the simulated and experimented results, and therefore the authors would like to propose relevant recommendations for the improvement of Geant4-DNA.
References
[1] S. Incerti, et al. (totally 13 authors), The Geant4-DNA Project, Int. J. Model. Simul. Sci. Comput. 01 (2010) 157.
[2] R. Norarata, N. Semsang, S. Anuntalabhochai, L.D. Yu, Very low-energy and low-fluence ion beam bombardment of naked plasmid DNA, Nucl. Instr. Meth. B 267 (2009) 1650-1653.
[3] P. Thopan, L.D. Yu, U. Tippawan, Critical low-energy Ar-ion beam conditions to induce direct DNA double strand break, Surf. Coat. Technol. 306 (2016) 313-318.
[4] C. Ngaojampa, P. Nimmanpipug, L.D. Yu, S. Anuntalabhochai, V.S. Lee, Molecular simulations of ultra-low-energy nitrogen ion bombardment of A-DNA in vacuum, Journal of Molecular Graphics and Modelling 28 (2010) 533-539.
[5] Z.L. Yu, Introduction to Ion Beam Biotechnology, translated by L.D. Yu, T. Vilaithong and I. Brown, Springer, New York, 2006.
[6] C. Ngaojampa, P. Nimmanpipug, L.D. Yu, S. Anuntalabhochai, V.S. Lee, Combined quantum-mechanics/molecular-mechanics dynamics simulation of A-DNA double strands irradiated by ultra-low-energy carbon ions, Nucl. Instr. Meth. B 269 (2011) 504-510.
Reported by
Dr. L. D. Yu, Senior Research Fellow, Thailand Center of Excellence in Physics (email: yuld@thep-center.org), based on a very recent publication of a paper entitled Geant4-DNA simulation of radiation effects in DNA on strand breaks from ultra-low-energy particles, Nucl. Instr. Meth. B 475 (2020) 77-83, by P. Nimmanpipug*, V.S. Lee, P. Kruanopparatana, and L.D. Yu* (* corresponding authors).